Projects
Causal Mediation Analysis of Carbohydrate Intake and Insulin on Postprandial Glycemia
Mobile health (mHealth) which uses digital devices to collect real-time data and deliver health care interventions, has emerged as accessible tools to support various health behaviors, including continuous glucose monitoring, physical activity tracking, and mental health management. Careful analysis of mHealth data has the potential to provide improved and individualized care for patients. However, the high frequency, longitudinal scope, and multi-modal nature of mHealth data introduce unique methodological challenges, complicating analysis and the extraction of actionable insights. This research aims to integrate statistical theory with deep learning techniques to address these challenges and ultimately provide practitioners with precise and interpretable results.
Machine Learning for Multimodal Modeling and Early Detection of Alzheimer’s Disease
This research aims to develop interpretable and generalizable machine learning models that integrate neuroimaging, biomarker, and cognitive data to enhance the early detection and understanding of Alzheimer’s disease (AD). The first component focuses on modeling redundancy in diffusion-weighted imaging (DWI) metrics by utilizing statistical and unsupervised learning techniques to identify an optimized set of diffusion metrics. A second project expands early detection efforts by constructing a temporal event-based model to identify the order of decline across multiple cognitive domains during AD progression. Finally, a third project aims to predict regional tau PET burden using more accessible data sources such as structural MRI, DWI, and plasma biomarkers.
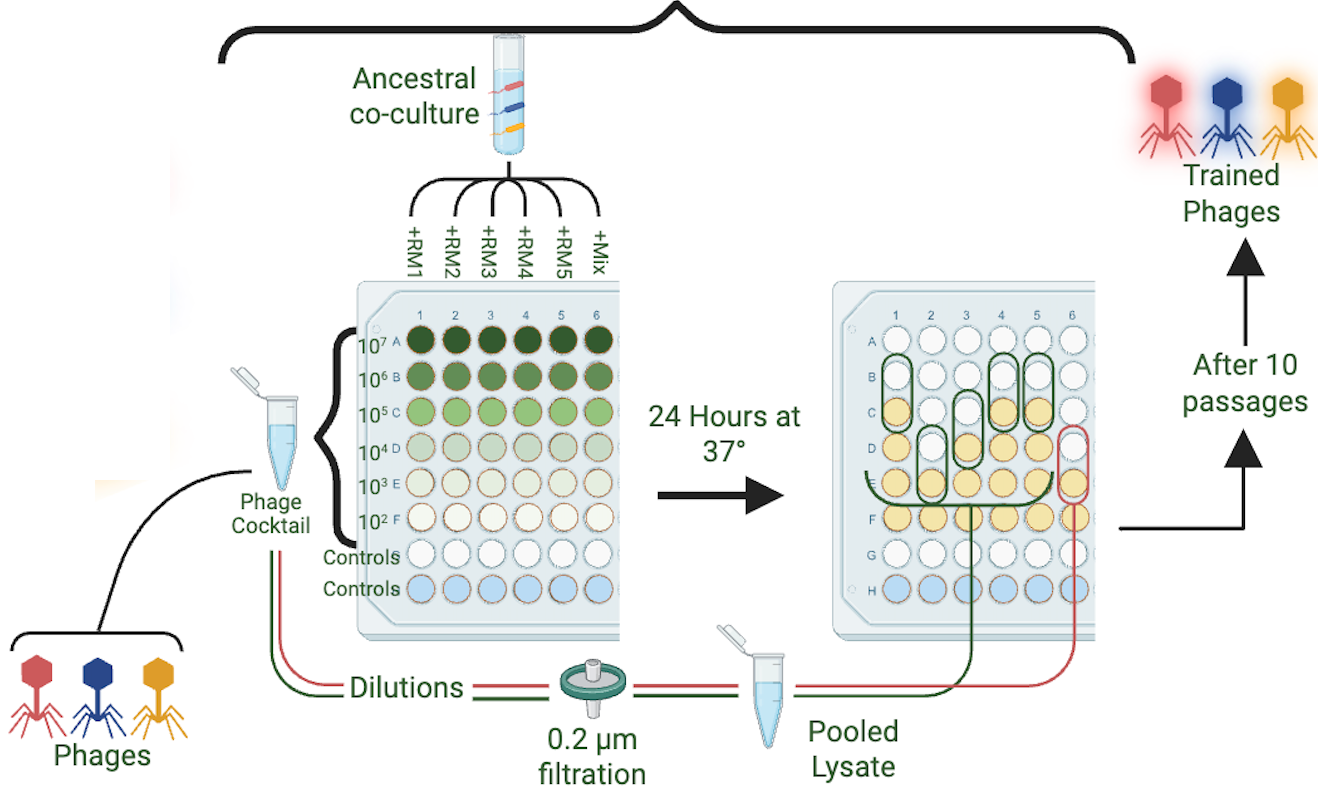
Overcoming Antibiotic Resistance through Data-Driven Phage Cocktail Design for Evolution-Proof Therapies
This research addresses the antimicrobial resistance crisis through the design of advanced therapeutic phage cocktails. We combine statistical modeling with directed evolution to design and assess phage cocktails with broad host ranges and minimized likelihood of bacterial resistance. By combining genetic changes in bacteria and phages with phenotypic outcomes like fitness, host range, and resistance suppression into a framework called CAPE (Cocktail Analysis and Predicted Evolution), this research aims to transform phage cocktail design from trial-and-error to a rigorous, evolution-informed process. Ultimately, bring phage therapy into the 21st century, and use nature’s perfect predator to tackle the antibiotic resistance crisis head-on.
Joint False Discovery Rate Control for Correlated High-Dimensional Genomic Data
This research focuses on developing and applying statistical methods to improve inference from high-dimensional biological data. The goal is to develop statistical models that enable more powerful procedures for multiple testing in genomics, with an overarching aim of creating robust methods that account for dependencies across data sources. Typically, in large-scale genomics studies thousands of hypotheses are tested simultaneously, often across related conditions. Standard false discovery rate (FDR) procedures typically treat each condition independently, which can reduce power and obscure true signals. This research develops a joint FDR framework that explicitly models correlation between test statistics across conditions. Simulations and applications to mouse astrocyte gene expression data show that the joint approach improves sensitivity while maintaining FDR control, uncovering significant genes missed by univariate methods.